Basic file loading operations
The core of all the data-loading operations is the FileManager class defined in the read_input submodule. The FileManager class can load both .DTA and .mpt files generated respectively by the GAMRY and BIOLOGIC potentiostats. The class provides a simple interface to automatically parse all the loaded data and hold them in memory in the form of bytestreams. To show how this object can be operated let us consider the case of a .mpt file that, for the purposes of this example, is provided in the documentation folder. Let us look at the content of such file:
with open("./example.mpt", 'r') as file:
for line in file:
print(line.rstrip('\n'))
...
Acquisition started on : 25/12/2022 13:00:00
...
Number of loops : 2
Loop 0 from point number 0 to 5
Loop 1 from point number 6 to 11
mode ox/red error control changes time/s control/V/mA Ewe/V I/mA dq/mA.h (Q-Qo)/mA.h Q charge/discharge/mA.h Ece/V P/W Q discharge/mA.h Q charge/mA.h Capacity/mA.h control/V control/mA Ewe-Ece/V
1 1 0 1 1,000000000000000E+002 8,0000000E+002 1,0000000E+000 8,0000000E+002 4,000000000000000E+000 1,100000000000000E+003 1,100000000000000E+003 3,0000000E-006 1,0000000E+000 0,000000000000000E+000 1,000000000000000E+003 1,000000000000000E+003 0,0000000E+000 8,0000000E+002 1,0000000E+000
1 1 0 1 1,010000000000000E+002 8,0000000E+002 1,1000000E+000 8,0000000E+002 4,000000000000000E+000 1,100000000000000E+003 1,100000000000000E+003 3,0000000E-006 1,0000000E+000 0,000000000000000E+000 1,000000000000000E+003 1,000000000000000E+003 0,0000000E+000 8,0000000E+002 1,0000000E+000
1 1 0 1 1,020000000000000E+002 8,0000000E+002 1,2000000E+000 8,0000000E+002 4,000000000000000E+000 1,100000000000000E+003 1,100000000000000E+003 3,0000000E-006 1,0000000E+000 0,000000000000000E+000 1,000000000000000E+003 1,000000000000000E+003 0,0000000E+000 8,0000000E+002 1,0000000E+000
1 0 0 0 1,030000000000000E+002 -8,0000000E+002 9,0000000E-001 -8,0000000E+002 -4,000000000000000E-005 1,100000000000000E+003 -4,000000000000000E-005 -8,0000000E-004 -4,0000000E-001 5,000000000000000E-005 0,000000000000000E+000 5,000000000000000E-005 0,0000000E+000 -8,0000000E+002 6,0000000E-001
1 0 0 0 1,040000000000000E+002 -8,0000000E+002 8,5000000E-001 -8,0000000E+002 -4,000000000000000E+000 1,100000000000000E+003 -4,000000000000000E+000 -8,0000000E-005 -4,0000000E-001 5,000000000000000E+000 0,000000000000000E+000 5,000000000000000E+000 0,0000000E+000 -8,0000000E+002 6,0000000E-001
1 0 0 0 1,050000000000000E+002 -8,0000000E+002 8,2000000E-001 -8,0000000E+002 -4,000000000000000E+000 1,100000000000000E+003 -4,000000000000000E+000 -8,0000000E-004 -4,0000000E-001 5,000000000000000E+000 0,000000000000000E+000 5,000000000000000E+000 0,0000000E+000 -8,0000000E+002 6,0000000E-001
1 1 0 1 1,060000000000000E+002 8,0000000E+002 1,0000000E+000 8,0000000E+002 4,000000000000000E+000 1,100000000000000E+003 1,100000000000000E+003 3,0000000E-006 1,0000000E+000 0,000000000000000E+000 1,000000000000000E+003 1,000000000000000E+003 0,0000000E+000 8,0000000E+002 1,0000000E+000
1 1 0 1 1,070000000000000E+002 8,0000000E+002 1,0500000E+000 8,0000000E+002 4,000000000000000E+000 1,100000000000000E+003 1,100000000000000E+003 3,0000000E-006 1,0000000E+000 0,000000000000000E+000 1,000000000000000E+003 1,000000000000000E+003 0,0000000E+000 8,0000000E+002 1,0000000E+000
1 1 0 1 1,080000000000000E+002 8,0000000E+002 1,1500000E+000 8,0000000E+002 4,000000000000000E+000 1,100000000000000E+003 1,100000000000000E+003 3,0000000E-006 1,0000000E+000 0,000000000000000E+000 1,000000000000000E+003 1,000000000000000E+003 0,0000000E+000 8,0000000E+002 1,0000000E+000
1 0 0 0 1,090000000000000E+002 -8,0000000E+002 8,9000000E-001 -8,0000000E+002 -4,000000000000000E-005 1,100000000000000E+003 -4,000000000000000E-005 -8,0000000E-004 -4,0000000E-001 5,000000000000000E-005 0,000000000000000E+000 5,000000000000000E-005 0,0000000E+000 -8,0000000E+002 6,0000000E-001
1 0 0 0 1,100000000000000E+002 -8,0000000E+002 8,4000000E-001 -8,0000000E+002 -4,000000000000000E+000 1,100000000000000E+003 -4,000000000000000E+000 -8,0000000E-005 -4,0000000E-001 5,000000000000000E+000 0,000000000000000E+000 5,000000000000000E+000 0,0000000E+000 -8,0000000E+002 6,0000000E-001
1 0 0 0 1,110000000000000E+002 -8,0000000E+002 8,0000000E-001 -8,0000000E+002 -4,000000000000000E+000 1,100000000000000E+003 -4,000000000000000E+000 -8,0000000E-004 -4,0000000E-001 5,000000000000000E+000 0,000000000000000E+000 5,000000000000000E+000 0,0000000E+000 -8,0000000E+002 6,0000000E-001
As it can be seen, the provided file describes: two full charge/discharge cycles, each of wich composed by two halfcycle, each of which composed by 3 time-steps.
To load this .mpt file using the FileManager class, the following procedure can be used:
# Loading the FileManager class from the cellcycling.read_input module
from echemsuite.cellcycling.read_input import FileManager
# Creating an instance of the FileManager class
manager = FileManager(verbose=False)
# Loading all the .mpt file in the current folder
manager.fetch_from_folder("./", extension=".mpt")
After the call to the fetch_from_folder function, all the data has been loaded in the class and parsed in single independent half-cycles.
Loading files manually
Please notice how the user can also manually specify a list of files to be loaded by using the function fetch_files according to the syntax:
manager = FileManager()
manager.fetch_files(["./example.mpt"])
Please notice how in this case the file extension is automatically detected from the filenames contained in the list. As such the extension must be available and omogeneous between the various files in the list.
Considering that in the previous example we have loaded a single .mpt file, the half-cycle ordering is unambiguos and the parsed data can immediately be converted into the corresponding Cycle and CellCycling objects by using the built-in get_cycles and get_cellcycling class methods:
# Obtain the cellcycling object from the manager
cellcycling = manager.get_cellcycling()
print(cellcycling)
# Obtain the list of cycle objects from the manager
cycle_list = manager.get_cycles()
print(cycle_list[0])
<echemsuite.cellcycling.cycles.CellCycling at 0x7ff05ea42e80>
├─ timestamp: 2022-12-25 13:01:40
├─ total number of cycles: 2
├─ number of visible cycles: 2
└─ reference cycle: 0
<echemsuite.cellcycling.cycles.Cycle at 0x7ff0989842b0>
├─ number: 0
├─ halfcycles: Both charge and discharge
└─ hidden: False
If the cell-cycling data is instead loaded from a set of .DTA files, either complete or partial, the ordering of the half-cycles is not strictly encoded by the data available to the FileManager and, as such, user intervention may be needed. The FileManager will try to automatically generate a suggested ordering of the half-cycle files based on their timestamp and type. The suggested ordering can be obtained by calling the memeber function suggest_ordering that will return a list of lists containing the name of the loaded files. Each list-type entry represents a half-cycle level; lists with more than one element will represent partial half-cycle files to be merged. As an example consider the followiing list:
[["Charge_1.DTA"], ["Charge_2.DTA", "Charge_2b.DTA"], ["Charge_3.DTA"]]
this list represents an ordering that will generate 3 half-cycles in which the middle one will be generated from the merging of Charge_2.DTA and Charge_2b.DTA. The user can either accept the automatically generated ordering or can manually specify its own. The selected ordering can then be applyed by passing it explicitly to the get_cycles and get_cellcycling functions as the keyworded argument custom_order.
To better show how this can be done, let us consider the following example in which two partial .DTA halfcycle charge files are provided, together with a complete discharge halfcycle file, to the file-manager.
import os
import matplotlib.pyplot as plt
from echemsuite.cellcycling.read_input import FileManager
from echemsuite.cellcycling.cycles import Cycle
# Define the path to the folder holding the files
folder = "./example_partial_DTA"
# Print a list of the available files
print(f"Available files: {', '.join([f for f in os.listdir(folder)])}\n")
# Load and parse the .DTA files contained in the "my_folder" directory
manager = FileManager()
manager.fetch_from_folder(folder, ".DTA")
# Print a list of the loaded halfcycles
print("Loaded halfcycles datasets:")
for name, obj in manager.halfcycles.items():
print(f"{obj.timestamp}: ({obj.halfcycle_type}) {name}")
print("")
# Print the suggested ordering of the loaded-halfcycles
print("Ordering suggested by the FileManager object:")
print(manager.suggest_ordering())
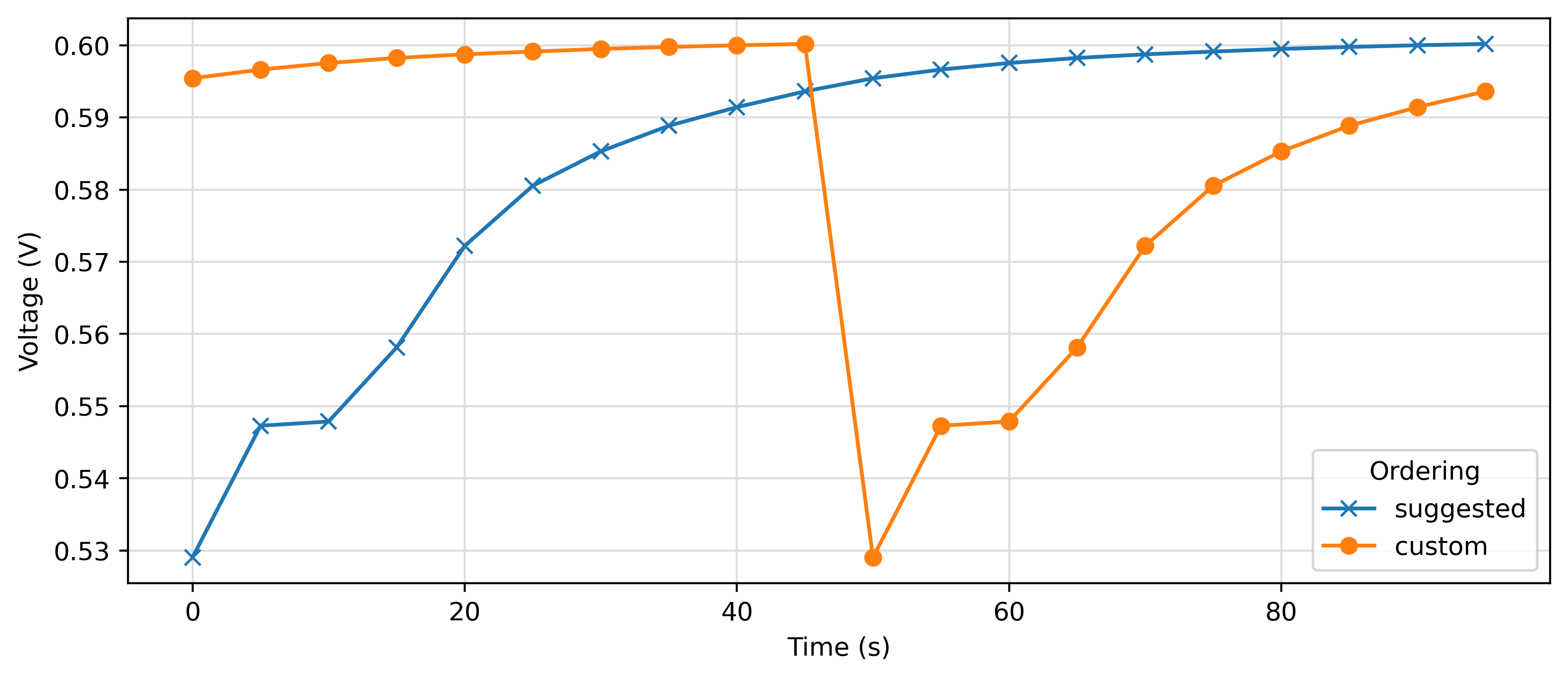
# Plotting the result obtained by concatenating the data according to the suggested ordering
# and according to a user-specified ordering
suggested = manager.get_cellcycling()
custom = manager.get_cellcycling(custom_order=[['charge_1b.DTA', 'charge_1.DTA'], ['discharge_1.DTA']] )
fig = plt.figure(figsize=(10, 4), dpi=400)
plt.plot(suggested[0].charge.time, suggested[0].charge.voltage, label="suggested", marker="x")
plt.plot(custom[0].charge.time, custom[0].charge.voltage, label="custom", marker="o")
plt.xlabel("Time (s)")
plt.ylabel("Voltage (V)")
plt.grid(which="major", c="#DDDDDD")
plt.legend(title="Ordering")
Available files: charge_1b.DTA, charge_1.DTA, discharge_1.DTA
Loaded halfcycles datasets:
2022-07-23 20:28:33: (charge) charge_1b.DTA
2022-07-23 20:25:33: (charge) charge_1.DTA
2022-07-24 00:23:03: (discharge) discharge_1.DTA
Ordering suggested by the FileManager object:
[['charge_1.DTA', 'charge_1b.DTA'], ['discharge_1.DTA']]
<matplotlib.legend.Legend at 0x7ff05cc34f40>
Loading data from a BytesIO bytestream
The read_input sub-module has been developed to also operate in a web-application environment minimizing the I/O calls done on local copies of the loaded files. For this reason, all the loaded files are immediately converted into binary bytestreams and saved in a bytestrem dictionary saved in memory. By taking advantage of this behavior, a parametrized FileManager object can also be generated by directly setting the bytestream buffer via the provided setter.
To show how this can be done let us, first of all, define a BytesIO version of the .mpt file considered before. This can easily be done with few lines of code:
# Load the BytesIO stream object from the io module
from io import BytesIO
# Open the file and load it as a bytes stream
my_bytestream = None
with open("./example.mpt", 'rb') as binary_file:
my_bytestream = BytesIO(binary_file.read())
The loaded stream can now be used to directly initialize the FileManager class according to:
from echemsuite.cellcycling.read_input import Instrument
# Create a FileManager instance
manager = FileManager()
# Set the bytestream dictionary using the loaded data
manager.bytestreams = {"example.mpt" : my_bytestream}
manager._instrument = Instrument.BIOLOGIC
# Call the parse function
manager.parse()
Please observe how the FileManager class has no access to the file extension (the label provided here as a dictionary key was used only for clarity) and, as a direct consequence, does not know the instrument type. This requires the user to manually set the _instrument variable. Furthermore, unlike the previous example in which the call to the parse function was avoided thanks to the autoparse mode of the fetch_from_folder function, this time the call to the parse function must be made explicitly.